Frequently Asked Questions
- Is all of the data in this database available for free?
- How do I download CEL files of the microarray experiments ?
- How does RefDIC integrate microarray and proteome data on this database?
- What is the difference between MAS5- and gcRMA-processed data ?
Yes. All of the data in RefDIC is freely available as long as it is not used for commercial purposes. If you belong to a commercial entitiy, please contact us.
Affymetrix GeneChip CEL files of each sample can be downloaded from "Data sets" section. First, search a title of your interested sample by 'Search' or 'Browser'. You can find a link to "Microarray data" page at the foot of "Attribute" page.
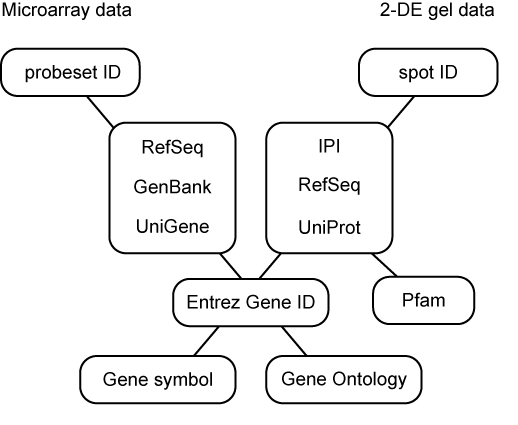
To do this, we took advantage of the Entrez Gene database at NCBI because it provides solid references for various lines of genomic information including transcript and protein sequences, with a unique, stable and traceable gene identifier (Gene ID) as a central hub to integrate these data on RefDIC. A schematic diagram of the relationship between probe set identifiers of microarray data and spot identifiers of 2-DE gel based proteome experiments on RefDIC is shown below.
MAS5, originally developed by Affymetrix Inc., is a basic algorithm to estimate the expression level from the signal intensities of probes in a probe set. On the other hand, gcRMA is an improved version of RMA (Robust MultiChip Averaging), developed by Irizarry et al. (2003), GC-content of each probe sequence is taken for the background correction, and this algorithm is superior than MAS5 in terms of the ability to detect differentially expressed genes. As several groups evaluated and discussed the reliability of these algorithms, please refer to these articles for more details (Ref. 1, Ref. 2).
